library(cmdstanr)
library(dplyr)
library(ggplot2)Nominal Regression in STAN
I was talking to a colleague about modeling nominal outcomes in STAN, and wrote up this example. Just put it here in case it’s helpful for anyone (probably myself in the future). This is based on an example I made for a course, where you can find the brms code for nominal regression. Please also check out the Multi-logit regression session on the Stan User’s guide.
Load Packages
Check out this paper: https://journals.sagepub.com/doi/full/10.1177/2515245918823199
stemcell <- read.csv("https://osf.io/vxw73/download")stemcell |>
ggplot(aes(x = rating)) +
geom_bar() +
facet_wrap(~ gender)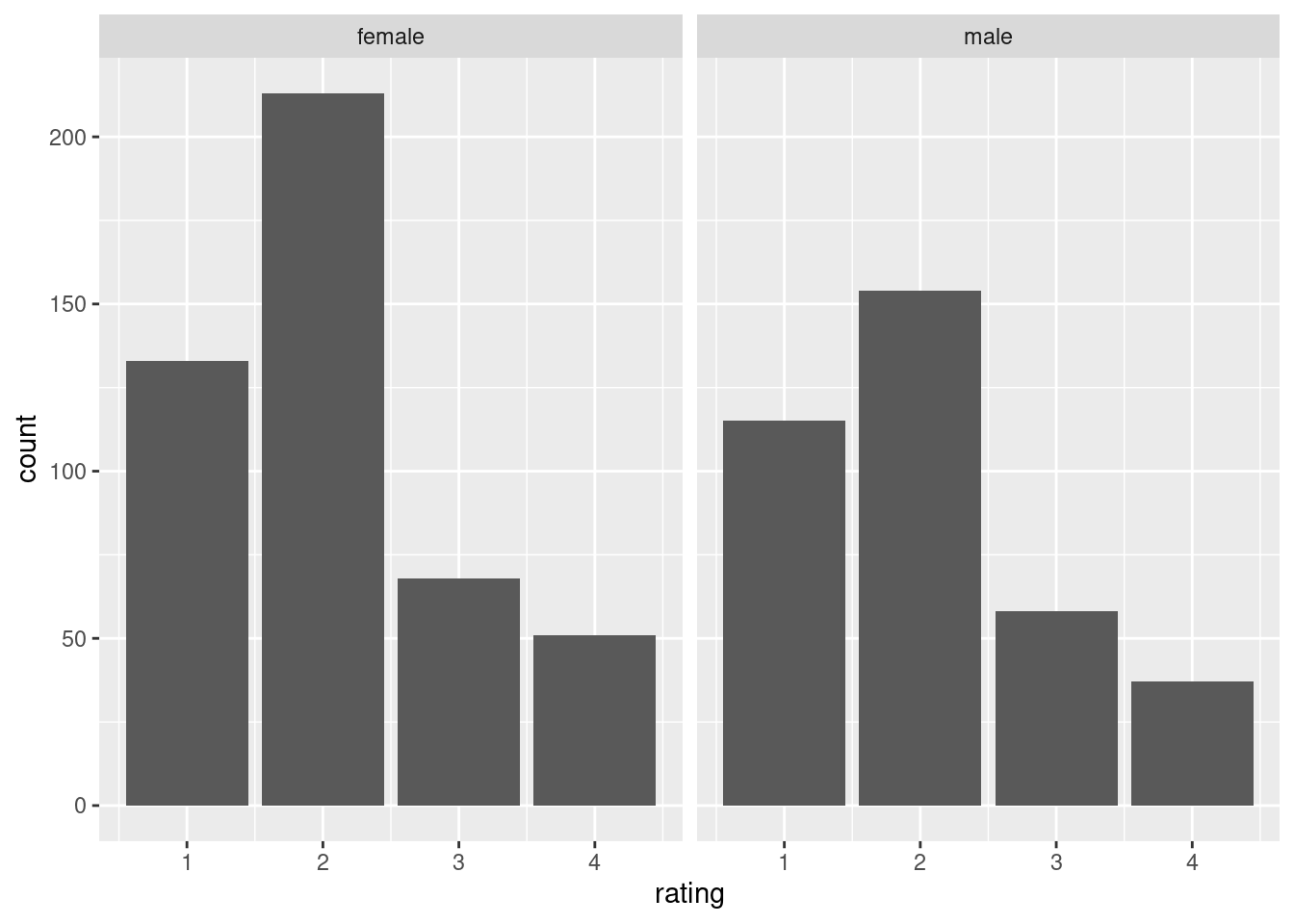
https://www.thearda.com/archive/files/Codebooks/GSS2006_CB.asp
The outcome is attitude towards stem cells research, and the predictor is gender.
Recently, there has been controversy over whether the government should provide any funds at all for scientific research that uses stem cells taken from human embryos. Would you say the government . . .
- 1 = Definitely, should fund such research
- 2 = Probably should fund such research
- 3 = Probably should not fund such research
- 4 = Definitely should not fund such research
Nominal Logistic Regression
Ordinal regression is a special case of nominal regression with the proportional odds assumption.
Model
\[\begin{align} \text{rating}_i & \sim \mathrm{Categorical}(\pi^1_{i}, \pi^2_{i}, \pi^3_{i}, \pi^4_{i}) \\ \pi^1_{i} & = \frac{1}{\exp(\eta^2_{i}) + \exp(\eta^3_{i}) + \exp(\eta^4_{i}) + 1} \\ \pi^2_{i} & = \frac{\exp(\eta^2_{i})}{\exp(\eta^2_{i}) + \exp(\eta^3_{i}) + \exp(\eta^4_{i}) + 1} \\ \pi^3_{i} & = \frac{\exp(\eta^3_{i})}{\exp(\eta^2_{i}) + \exp(\eta^3_{i}) + \exp(\eta^4_{i}) + 1} \\ \pi^4_{i} & = \frac{\exp(\eta^4_{i})}{\exp(\eta^2_{i}) + \exp(\eta^3_{i}) + \exp(\eta^4_{i}) + 1} \\ \eta^2_{i} & = \beta^2_{0} + \beta^2_{1} \text{male}_{i} \\ \eta^3_{i} & = \beta^3_{0} + \beta^3_{1} \text{male}_{i} \\ \eta^4_{i} & = \beta^4_{0} + \beta^4_{1} \text{male}_{i} \\ \end{align}\]
mod <- cmdstan_model("nominal_reg.stan")
mod//
// This Stan program defines a nominal regression model.
//
// It is based on
// https://mc-stan.org/docs/stan-users-guide/multi-logit.html
//
// The input data is a vector 'y' of length 'N'.
data {
int<lower=0> K; // number of response categories
int<lower=0> N; // number of observations (data rows)
int<lower=0> D; // number of predictors
array[N] int<lower=1, upper=K> y; // response vector
matrix[N, D] x; // predictor matrix
}
transformed data {
vector[D] zeros = rep_vector(0, D);
}
// The parameters accepted by the model.
parameters {
vector[K - 1] b0_raw; // intercept for second to last categories
matrix[D, K - 1] beta_raw;
}
// The model to be estimated.
model {
// Add zeros for reference category
vector[K] b0 = append_row(0, b0_raw);
matrix[D, K] beta = append_col(zeros, beta_raw);
to_vector(beta_raw) ~ normal(0, 5);
y ~ categorical_logit_glm(x, b0, beta);
}stan_dat <- with(stemcell,
list(K = n_distinct(rating),
N = length(rating),
D = 1,
y = rating,
x = matrix(as.integer(gender == "male")))
)# Draw samples
fit <- mod$sample(data = stan_dat, seed = 123, chains = 4,
parallel_chains = 2, refresh = 500,
iter_sampling = 2000, iter_warmup = 2000)fit$summary() |>
knitr::kable()| variable | mean | median | sd | mad | q5 | q95 | rhat | ess_bulk | ess_tail |
|---|---|---|---|---|---|---|---|---|---|
| lp__ | -1035.3043988 | -1034.9800000 | 1.7372773 | 1.5863820 | -1038.6200000 | -1033.1495000 | 1.000844 | 3578.616 | 5343.796 |
| b0_raw[1] | 0.4697455 | 0.4689280 | 0.1105168 | 0.1110905 | 0.2891216 | 0.6530174 | 1.001056 | 4017.342 | 5352.187 |
| b0_raw[2] | -0.6797744 | -0.6782500 | 0.1495865 | 0.1485951 | -0.9299347 | -0.4383052 | 1.000235 | 4383.623 | 5447.054 |
| b0_raw[3] | -0.9695071 | -0.9694105 | 0.1636312 | 0.1642639 | -1.2425745 | -0.7015362 | 1.000692 | 4591.053 | 5376.113 |
| beta_raw[1,1] | -0.1747948 | -0.1736585 | 0.1666419 | 0.1655399 | -0.4543450 | 0.1009604 | 1.001103 | 4012.471 | 5258.483 |
| beta_raw[1,2] | -0.0074333 | -0.0060478 | 0.2177309 | 0.2196757 | -0.3693494 | 0.3494878 | 1.000258 | 4256.543 | 5283.208 |
| beta_raw[1,3] | -0.1722835 | -0.1734355 | 0.2474491 | 0.2485277 | -0.5740370 | 0.2324758 | 1.000817 | 4722.193 | 5370.922 |
Compare to brms:
library(brms)
brm(rating ~ gender, data = stemcell, family = categorical(link = "logit"),
file = "mlogit") Family: categorical
Links: mu2 = logit; mu3 = logit; mu4 = logit
Formula: rating ~ gender
Data: stemcell (Number of observations: 829)
Draws: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 4000
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
mu2_Intercept 0.47 0.11 0.26 0.69 1.00 3990 3108
mu3_Intercept -0.67 0.15 -0.97 -0.39 1.00 3809 3130
mu4_Intercept -0.96 0.17 -1.29 -0.63 1.00 4027 3031
mu2_gendermale -0.18 0.17 -0.51 0.15 1.00 4272 2829
mu3_gendermale -0.01 0.22 -0.43 0.41 1.00 4141 2902
mu4_gendermale -0.17 0.25 -0.67 0.34 1.00 3922 2937
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).The estimates are pretty much the same.